A Guide to Installing TorchDrug 0.1.0 (CPU) and Using with Jupyter Lab
Published:
TorchDrug, a platform for drug discovery with PyTorch, was just released. It was developed at MILA with Jian Tang as the principal investigator and launched on GitHub by Zhaocheng Zhu (full team here).
Background and Outline
I was eager to dig in but found that, as with many brand new deep learning frameworks, the installation did not go smoothly at first. Maybe it’s because I’m using the CPU installation. I am using Linux so I’m not sure whether Windows would be harder or easier. I ultimately got it working and want to document the procedure in case it could be helpful to others. Also, if you are having trouble with a GPU installation, this may still be relevant, as following the same basic steps could be helpful for you.
Outline:
- Installation instructions. I have Ubuntu Linux 20.04, 64 bit, but Mac should work similarly.
- Testing your implementation
- Additional details/explanation
I’m sure the devs will streamline the installation, but this could help the community get up and running for now!
Installation Steps
I assume use of a Conda environment with pip installed inside of that env. The following commands are executed in a terminal. (I generally try to avoid the practice of mixing pip and conda calls, but sometimes it’s hard to avoid)
Create an Anaconda environment:
conda create -n td_test python=3.8
conda activate td_test
Install PyTorch 1.8.1 (for CPU in my case)
pip3 install torch==1.8.1+cpu torchvision==0.9.1+cpu torchaudio==0.8.1 -f https://download.pytorch.org/whl/lts/1.8/torch_lts.html
Install Torch Scatter, using the specific version for PyTorch 1.8.0/1.8.1 (reference: pytorch_scatter github)
pip install torch-scatter -f https://pytorch-geometric.com/whl/torch-1.8.0+cpu.html
Install rdkit to work with molecules
conda install -c conda-forge rdkit
Build from source
mkdir ~/torch_drug_test
cd ~/torch_drug_test
git clone https://github.com/DeepGraphLearning/torchdrug
cd torchdrug
# Install the other packages in requirements.txt:
Make sure you open requirements.txt and install the other packages (do not reinstall torch or torch-scatter this way). I used pip.
Finally,
python setup.py install
For Jupyter Lab to work, follow these instructions (reference)
conda install -c conda-forge jupyterlab
conda install ipykernel
ipython kernel install --user
Testing Installation
To test the install, we can try the code in the Quick Start Guide.
A Note about CPU Installations
When I first wrote this post, I had to manually fix a small bug in the TorchDrug code. It has been fixed by the devs, which I confirmed myself using a fresh install. The devs have also confirmed that they do plan to support CPU installations. (You can see my thread with the devs here.) You should be able to skip the manual fix below, which I include for reference.
There is a cuda call that can confuse CPU installations. I’m not sure what the developers are planning with regards to CPU support, and I have asked them. For now, let us proceed with a quick fix. I’m sure the community will find a solution and the team will release a patch.
Open
~/torch_drug_test/torchdrug/torchdrug/core/meter.py
and comment-out line 84
torch.cuda.reset_peak_memory_stats()
or use an if statement to turn it off when no device is available.
Run in Jupyter Lab
What follows is from their Quick Start Guide; I just removed another reference to cuda.
import torch
import torchdrug as td
from torchdrug import data, datasets, core, models, tasks
%matplotlib inline
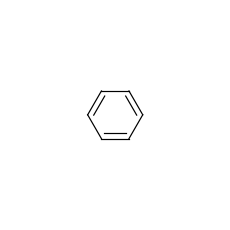
mol = data.Molecule.from_smiles("C1=CC=CC=C1")
mol.visualize()
print(mol.node_feature.shape)
print(mol.edge_feature.shape)
torch.Size([6, 69])
torch.Size([12, 19])
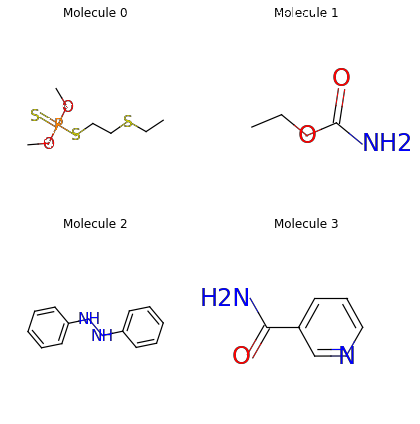
smiles_list = ["CCSCCSP(=S)(OC)OC", "CCOC(=O)N",
"N(Nc1ccccc1)c2ccccc2", "NC(=O)c1cccnc1"]
mols = data.PackedMolecule.from_smiles(smiles_list)
mols.visualize()
# mols = mols.cuda()
print(mols)
PackedMolecule(batch_size=4, num_nodes=[12, 6, 14, 9], num_edges=[22, 10, 30, 18], num_relation=4)
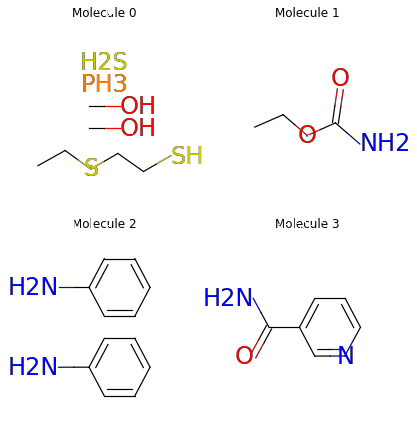
node_in, node_out, bond_type = mols.edge_list.t()
edge_mask = (mols.atom_type[node_in] == td.CARBON) | \
(mols.atom_type[node_out] == td.CARBON)
mols = mols.edge_mask(edge_mask)
mols.visualize()
dataset = datasets.ClinTox("~/molecule-datasets/")
lengths = [int(0.8 * len(dataset)), int(0.1 * len(dataset))]
lengths += [len(dataset) - sum(lengths)]
train_set, valid_set, test_set = torch.utils.data.random_split(dataset, lengths)
Loading /home/murph213/molecule-datasets/clintox.csv: 100%|██████████| 1485/1485 [00:00<00:00, 40417.52it/s]
Constructing molecules from SMILES: 0%| | 0/1484 [00:00<?, ?it/s]/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `*`
warnings.warn("Unknown value `%s`" % x)
RDKit ERROR: [18:54:20] Explicit valence for atom # 0 N, 5, is greater than permitted
/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Tc`
warnings.warn("Unknown value `%s`" % x)
/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Fe`
warnings.warn("Unknown value `%s`" % x)
/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Al`
warnings.warn("Unknown value `%s`" % x)
Constructing molecules from SMILES: 7%|▋ | 110/1484 [00:00<00:02, 487.53it/s]/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Ca`
warnings.warn("Unknown value `%s`" % x)
Constructing molecules from SMILES: 11%|█ | 160/1484 [00:00<00:03, 440.73it/s]/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Pt`
warnings.warn("Unknown value `%s`" % x)
Constructing molecules from SMILES: 20%|█▉ | 295/1484 [00:00<00:02, 423.90it/s]RDKit ERROR: [18:54:21] Can't kekulize mol. Unkekulized atoms: 9
RDKit ERROR:
/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Bi`
warnings.warn("Unknown value `%s`" % x)
Constructing molecules from SMILES: 66%|██████▌ | 973/1484 [00:02<00:01, 293.10it/s]RDKit ERROR: [18:54:23] Explicit valence for atom # 10 N, 4, is greater than permitted
RDKit ERROR: [18:54:23] Explicit valence for atom # 10 N, 4, is greater than permitted
Constructing molecules from SMILES: > 81%|████████▏ | 1209/1484 [00:03<00:00, 320.23it/s]RDKit ERROR: [18:54:24] Can't kekulize mol. Unkekulized atoms: 4
RDKit ERROR:
RDKit ERROR: [18:54:24] Can't kekulize mol. Unkekulized atoms: 4
RDKit ERROR:
Constructing molecules from SMILES: 90%|█████████ | 1336/1484 [00:04<00:00, 270.75it/s]/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Au`
warnings.warn("Unknown value `%s`" % x)
/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Tl`
warnings.warn("Unknown value `%s`" % x)
/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Cr`
warnings.warn("Unknown value `%s`" % x)
/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Mn`
warnings.warn("Unknown value `%s`" % x)
Constructing molecules from SMILES: 95%|█████████▌| 1410/1484 [00:04<00:00, 314.47it/s]/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Hg`
warnings.warn("Unknown value `%s`" % x)
Constructing molecules from SMILES: 99%|█████████▉| 1473/1484 [00:04<00:00, 297.11it/s]/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `As`
warnings.warn("Unknown value `%s`" % x)
/home/murph213/anaconda3/envs/torchdrug/lib/python3.8/site-packages/torchdrug-0.1.0-py3.8.egg/torchdrug/data/feature.py:37: UserWarning: Unknown value `Ti`
warnings.warn("Unknown value `%s`" % x)
Constructing molecules from SMILES: 100%|██████████| 1484/1484 [00:04<00:00, 321.71it/s]
model = models.GIN(input_dim=dataset.node_feature_dim,
hidden_dims=[256, 256, 256, 256],
short_cut=True, batch_norm=True, concat_hidden=True)
task = tasks.PropertyPrediction(model, task=dataset.tasks,
criterion="bce", metric=("auprc", "auroc"))
optimizer = torch.optim.Adam(task.parameters(), lr=1e-3)
solver = core.Engine(task, train_set, valid_set, test_set, optimizer,
batch_size=1024)
solver.train(num_epoch=3) # 3 epochs is enough to know whether the install is bug free
solver.evaluate("valid")
18:54:25 Preprocess training set
18:54:25 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
18:54:25 Epoch 0 begin
18:54:30 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
18:54:30 binary cross entropy: 4.37713
18:54:30 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
18:54:30 Epoch 0 end
18:54:30 duration: 5.55 secs
18:54:30 speed: 0.36 batch / sec
18:54:30 ETA: 11.10 secs
18:54:30 max GPU memory: 0.0 MiB
18:54:30 ------------------------------
18:54:30 average binary cross entropy: 3.57012
18:54:30 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
18:54:30 Epoch 1 begin
18:54:36 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
18:54:36 Epoch 1 end
18:54:36 duration: 5.42 secs
18:54:36 speed: 0.37 batch / sec
18:54:36 ETA: 5.49 secs
18:54:36 max GPU memory: 0.0 MiB
18:54:36 ------------------------------
18:54:36 average binary cross entropy: 2.05932
18:54:36 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
18:54:36 Epoch 2 begin
18:54:41 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
18:54:41 Epoch 2 end
18:54:41 duration: 5.57 secs
18:54:41 speed: 0.36 batch / sec
18:54:41 ETA: 0.00 secs
18:54:41 max GPU memory: 0.0 MiB
18:54:41 ------------------------------
18:54:41 average binary cross entropy: 4.61679
18:54:41 Evaluate on valid
18:54:41 >>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
18:54:41 auprc [CT_TOX]: 0.0562578
18:54:41 auprc [FDA_APPROVED]: 0.940434
18:54:41 auroc [CT_TOX]: 0.416264
18:54:41 auroc [FDA_APPROVED]: 0.408163
{'auprc [FDA_APPROVED]': tensor(0.9404),
'auprc [CT_TOX]': tensor(0.0563),
'auroc [FDA_APPROVED]': tensor(0.4082),
'auroc [CT_TOX]': tensor(0.4163)}
batch = data.graph_collate(valid_set[:8])
pred = task.predict(batch)
When this executes, we know that we have a functional installation. The RDKit error messages are likely a result of the data, not the installation.
Additional Details
When I tried to follow the installation instructions directly, OSErrors were thrown indicating problems with Torch Scatter. Also, when trying to use PyTorch 1.9, I ran into container_abcs errors. I arrived at these steps after trying a few things. Ultimately, I think the keys to success for this procedure were:
- Directly installing the correctly-versioned torch-scatter from the GitHub page, using
pip - Installing from source.
- Perhaps, the decision to use the Long Term Service version of PyTorch, 1.8.
Finally, I reiterate that I reached out to the devs and am sure a solution will be available. In the meantime, we can work with this installation to continue getting to know the framework and combing for any additional bugs!! AI in drug discovery has much potential, so it’s well worth pushing forward with this repo even as kinks get worked out!
Feel free to reach out with any questions.
